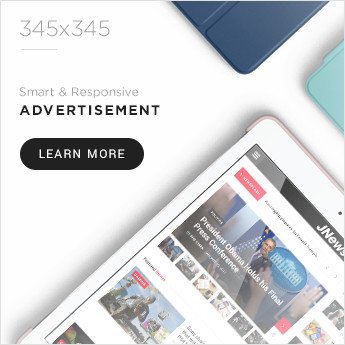
class BioPythonAIAgent:
def __init__(self, email="[email protected]"):
self.email = email
Entrez.email = email
self.sequences = {}
self.analysis_results = {}
self.alignments = {}
self.trees = {}
def fetch_sequence_from_ncbi(self, accession_id, db="nucleotide", rettype="fasta"):
try:
handle = Entrez.efetch(db=db, id=accession_id, rettype=rettype, retmode="text")
record = SeqIO.read(handle, "fasta")
handle.close()
self.sequences[accession_id] = record
return record
except Exception as e:
print(f"Error fetching sequence: {str(e)}")
return None
def create_sample_sequences(self):
covid_spike = "MFVFLVLLPLVSSQCVNLTTRTQLPPAYTNSFTRGVYYPDKVFRSSVLHSTQDLFLPFFSNVTWFHAIHVSGTNGTKRFDNPVLPFNDGVYFASTEKSNIIRGWIFGTTLDSKTQSLLIVNNATNVVIKVCEFQFCNDPFLGVYYHKNNKSWMESEFRVYSSANNCTFEYVSQPFLMDLEGKQGNFKNLREFVFKNIDGYFKIYSKHTPINLVRDLPQGFSALEPLVDLPIGINITRFQTLLALHRSYLTPGDSSSGWTAGAAAYYVGYLQPRTFLLKYNENGTITDAVDCALDPLSETKCTLKSFTVEKGIYQTSNFRVQPTESIVRFPNITNLCPFGEVFNATRFASVYAWNRKRISNCVADYSVLYNSASFSTFKCYGVSPTKLNDLCFTNVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDDFTGCVIAWNSNNLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTNGVGYQPYRVVVLSFELLHAPATVCGPKKSTNLVKNKCVNFNFNGLTGTGVLTESNKKFLPFQQFGRDIADTTDAVRDPQTLEILDITPCSFGGVSVITPGTNTSNQVAVLYQDVNCTEVPVAIHADQLTPTWRVYSTGSNVFQTRAGCLIGAEHVNNSYECDIPIGAGICASYQTQTNSPRRARSVASQSIIAYTMSLGAENSVAYSNNSIAIPTNFTISVTTEILPVSMTKTSVDCTMYICGDSTECSNLLLQYGSFCTQLNRALTGIAVEQDKNTQEVFAQVKQIYKTPPIKDFGGFNFSQILPDPSKPSKRSFIEDLLFNKVTLADAGFIKQYGDCLGDIAARDLICAQKFNGLTVLPPLLTDEMIAQYTSALLAGTITSGWTFGAGAALQIPFAMQMAYRFNGIGVTQNVLYENQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQLSSNFGAISSVLNDILSRLDKVEAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSECVLGQSKRVDFCGKGYHLMSFPQSAPHGVVFLHVTYVPAQEKNFTTAPAICHDGKAHFPREGVFVSNGTHWFVTQRNFYEPQIITTDNTFVSGNCDVVIGIVNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGINASVVNIQKEIDRLNEVAKNLNESLIDLQELGKYEQYIKWPWYIWLGFIAGLIAIVMVTIMLCCMTSCCSCLKGCCSCGSCCKFDEDDSEPVLKGVKLHYT"
human_insulin = "MALWMRLLPLLALLALWGPDPAAAFVNQHLCGSHLVEALYLVCGERGFFYTPKTRREAEDLQVGQVELGGGPGAGSLQPLALEGSLQKRGIVEQCCTSICSLYQLENYCN"
e_coli_16s = "AAATTGAAGAGTTTGATCATGGCTCAGATTGAACGCTGGCGGCAGGCCTAACACATGCAAGTCGAACGGTAACAGGAAGCAGCTTGCTGCTTTGCTGACGAGTGGCGGACGGGTGAGTAATGTCTGGGAAACTGCCTGATGGAGGGGGATAACTACTGGAAACGGTAGCTAATACCGCATAATGTCGCAAGACCAAAGAGGGGGACCTTCGGGCCTCTTGCCATCGGATGTGCCCAGATGGGATTAGCTAGTAGGTGGGGTAACGGCTCACCTAGGCGACGATCCCTAGCTGGTCTGAGAGGATGACCAGCCACACTGGAACTGAGACACGGTCCAGACTCCTACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCCATGCCGCGTGTATGAAGAAGGCCTTCGGGTTGTAAAGTACTTTCAGCGGGGAGGAAGGCGTTAAGGTTAATAACCTTGGCGATTGACGTTACCCGCAGAAGAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAATACGGAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGCACGCAGGCGGTCTGTCAAGTCGGATGTGAAATCCCCGGGCTCAACCTGGGAACTGCATCTGATACTGGCAAGCTTGAGTCTCGTAGAGGGGGGTAGAATTCCAGGTGTAGCGGTGAAATGCGTAGAGATCTGGAGGAATACCGGTGGCGAAGGCGGCCCCCTGGACAAAGACTGACGCTCAGGTGCGAAAGCGTGGGGAGCAAACA"
sample_sequences = [
("COVID_Spike", covid_spike, "SARS-CoV-2 Spike Protein"),
("Human_Insulin", human_insulin, "Human Insulin Precursor"),
("E_coli_16S", e_coli_16s, "E. coli 16S rRNA")
]
for seq_id, seq_str, desc in sample_sequences:
record = SeqRecord(Seq(seq_str), id=seq_id, description=desc)
self.sequences[seq_id] = record
return sample_sequences
def analyze_sequence(self, sequence_id=None, sequence=None):
if sequence_id and sequence_id in self.sequences:
seq_record = self.sequences[sequence_id]
seq = seq_record.seq
description = seq_record.description
elif sequence:
seq = Seq(sequence)
description = "Custom sequence"
else:
return None
analysis = {
'length': len(seq),
'composition': {}
}
for base in ['A', 'T', 'G', 'C']:
analysis['composition'][base] = seq.count(base)
if 'A' in analysis['composition'] and 'T' in analysis['composition']:
analysis['gc_content'] = round(gc_fraction(seq) * 100, 2)
try:
analysis['molecular_weight'] = round(molecular_weight(seq, seq_type="DNA"), 2)
except:
analysis['molecular_weight'] = len(seq) * 650
try:
if len(seq) % 3 == 0:
protein = seq.translate()
analysis['translation'] = str(protein)
analysis['stop_codons'] = protein.count('*')
if '*' not in str(protein)[:-1]:
prot_analysis = ProteinAnalysis(str(protein)[:-1])
analysis['protein_mw'] = round(prot_analysis.molecular_weight(), 2)
analysis['isoelectric_point'] = round(prot_analysis.isoelectric_point(), 2)
analysis['protein_composition'] = prot_analysis.get_amino_acids_percent()
except:
pass
key = sequence_id if sequence_id else "custom"
self.analysis_results[key] = analysis
return analysis
def visualize_composition(self, sequence_id):
if sequence_id not in self.analysis_results:
return
analysis = self.analysis_results[sequence_id]
fig = make_subplots(
rows=2, cols=2,
specs=[[{"type": "pie"}, {"type": "bar"}],
[{"colspan": 2}, None]],
subplot_titles=("Nucleotide Composition", "Base Count", "Sequence Properties")
)
labels = list(analysis['composition'].keys())
values = list(analysis['composition'].values())
fig.add_trace(
go.Pie(labels=labels, values=values, name="Composition"),
row=1, col=1
)
fig.add_trace(
go.Bar(x=labels, y=values, name="Count", marker_color=['red', 'blue', 'green', 'orange']),
row=1, col=2
)
properties = ['Length', 'GC%', 'MW (kDa)']
prop_values = [
analysis['length'],
analysis.get('gc_content', 0),
analysis.get('molecular_weight', 0) / 1000
]
fig.add_trace(
go.Scatter(x=properties, y=prop_values, mode="markers+lines",
marker=dict(size=10, color="purple"), name="Properties"),
row=2, col=1
)
fig.update_layout(
title=f"Comprehensive Analysis: {sequence_id}",
showlegend=False,
height=600
)
fig.show()
def perform_multiple_sequence_alignment(self, sequence_ids):
if len(sequence_ids) < 2:
return None
sequences = []
for seq_id in sequence_ids:
if seq_id in self.sequences:
sequences.append(self.sequences[seq_id])
if len(sequences) < 2:
return None
from Bio.Align import PairwiseAligner
aligner = PairwiseAligner()
aligner.match_score = 2
aligner.mismatch_score = -1
aligner.open_gap_score = -2
aligner.extend_gap_score = -0.5
alignments = []
for i in range(len(sequences)):
for j in range(i+1, len(sequences)):
alignment = aligner.align(sequences[i].seq, sequences[j].seq)[0]
alignments.append(alignment)
return alignments
def create_phylogenetic_tree(self, alignment_key=None, sequences=None):
if alignment_key and alignment_key in self.alignments:
alignment = self.alignments[alignment_key]
elif sequences:
records = []
for i, seq in enumerate(sequences):
record = SeqRecord(Seq(seq), id=f"seq_{i}")
records.append(record)
SeqIO.write(records, "temp.fasta", "fasta")
try:
clustalw_cline = ClustalwCommandline("clustalw2", infile="temp.fasta")
stdout, stderr = clustalw_cline()
alignment = AlignIO.read("temp.aln", "clustal")
os.remove("temp.fasta")
os.remove("temp.aln")
os.remove("temp.dnd")
except:
return None
else:
return None
calculator = DistanceCalculator('identity')
dm = calculator.get_distance(alignment)
constructor = DistanceTreeConstructor()
tree = constructor.upgma(dm)
tree_key = f"tree_{len(self.trees)}"
self.trees[tree_key] = tree
return tree
def visualize_tree(self, tree):
fig, ax = plt.subplots(figsize=(10, 6))
Phylo.draw(tree, axes=ax)
plt.title("Phylogenetic Tree")
plt.tight_layout()
plt.show()
def protein_structure_analysis(self, sequence_id):
if sequence_id not in self.sequences:
return None
seq = self.sequences[sequence_id].seq
try:
if len(seq) % 3 == 0:
protein = seq.translate()
if '*' not in str(protein)[:-1]:
prot_analysis = ProteinAnalysis(str(protein)[:-1])
structure_analysis = {
'molecular_weight': prot_analysis.molecular_weight(),
'isoelectric_point': prot_analysis.isoelectric_point(),
'amino_acid_percent': prot_analysis.get_amino_acids_percent(),
'secondary_structure': prot_analysis.secondary_structure_fraction(),
'flexibility': prot_analysis.flexibility(),
'gravy': prot_analysis.gravy()
}
return structure_analysis
except:
pass
return None
def comparative_analysis(self, sequence_ids):
results = []
for seq_id in sequence_ids:
if seq_id in self.analysis_results:
analysis = self.analysis_results[seq_id].copy()
analysis['sequence_id'] = seq_id
results.append(analysis)
df = pd.DataFrame(results)
if len(df) > 1:
fig = make_subplots(
rows=2, cols=2,
subplot_titles=("Length Comparison", "GC Content", "Molecular Weight", "Composition Heatmap")
)
fig.add_trace(
go.Bar(x=df['sequence_id'], y=df['length'], name="Length"),
row=1, col=1
)
if 'gc_content' in df.columns:
fig.add_trace(
go.Scatter(x=df['sequence_id'], y=df['gc_content'], mode="markers+lines", name="GC%"),
row=1, col=2
)
if 'molecular_weight' in df.columns:
fig.add_trace(
go.Bar(x=df['sequence_id'], y=df['molecular_weight'], name="MW"),
row=2, col=1
)
fig.update_layout(title="Comparative Sequence Analysis", height=600)
fig.show()
return df
def codon_usage_analysis(self, sequence_id):
if sequence_id not in self.sequences:
return None
seq = self.sequences[sequence_id].seq
if len(seq) % 3 != 0:
return None
codons = {}
for i in range(0, len(seq) - 2, 3):
codon = str(seq[i:i+3])
codons[codon] = codons.get(codon, 0) + 1
codon_df = pd.DataFrame(list(codons.items()), columns=['Codon', 'Count'])
codon_df = codon_df.sort_values('Count', ascending=False)
fig = px.bar(codon_df.head(20), x='Codon', y='Count',
title=f"Top 20 Codon Usage - {sequence_id}")
fig.show()
return codon_df
def motif_search(self, sequence_id, motif_pattern):
if sequence_id not in self.sequences:
return []
seq = str(self.sequences[sequence_id].seq)
positions = []
for i in range(len(seq) - len(motif_pattern) + 1):
if seq[i:i+len(motif_pattern)] == motif_pattern:
positions.append(i)
return positions
def gc_content_window(self, sequence_id, window_size=100):
if sequence_id not in self.sequences:
return None
seq = self.sequences[sequence_id].seq
gc_values = []
positions = []
for i in range(0, len(seq) - window_size + 1, window_size//4):
window = seq[i:i+window_size]
gc_values.append(gc_fraction(window) * 100)
positions.append(i + window_size//2)
fig = go.Figure()
fig.add_trace(go.Scatter(x=positions, y=gc_values, mode="lines+markers",
name=f'GC Content (window={window_size})'))
fig.update_layout(
title=f"GC Content Sliding Window Analysis - {sequence_id}",
xaxis_title="Position",
yaxis_title="GC Content (%)"
)
fig.show()
return positions, gc_values
def run_comprehensive_analysis(self, sequence_ids):
results = {}
for seq_id in sequence_ids:
if seq_id in self.sequences:
analysis = self.analyze_sequence(seq_id)
self.visualize_composition(seq_id)
gc_analysis = self.gc_content_window(seq_id)
codon_analysis = self.codon_usage_analysis(seq_id)
results[seq_id] = {
'basic_analysis': analysis,
'gc_window': gc_analysis,
'codon_usage': codon_analysis
}
if len(sequence_ids) > 1:
comparative_df = self.comparative_analysis(sequence_ids)
results['comparative'] = comparative_df
return results








![MarTech essential digital marketing tools [Infographic]](https://mgrowtech.com/wp-content/uploads/2025/08/MarTech-tools-wheel-graphic-and-copy-Aug2025-350x250.png)